Clinvar
The content on this website is based on ClinVar database version July 14, Simple ClinVar was developed to provide gene- and disease-wise summary statistic based on all available genetic variants from ClinVar, clinvar. How many missense variants are associated to heart disease? What are the top clinvar genes mutated clinvar Alzheimer?
NOTE: ClinVar is intended for use primarily by physicians and other professionals concerned with genetic disorders, by genetics researchers, and by advanced students in science and medicine. While the ClinVar database is open to all academic users, users seeking information about a personal medical or genetic condition are urged to consult with a qualified physician for diagnosis and for answers to personal questions. These tracks show the genomic positions of variants in the ClinVar database. ClinVar is a free, public archive of reports of the relationships among human variations and phenotypes, with supporting evidence. Because the ClinVar type no longer captures this information, any variation equal to or larger than 50 bp is now considered a CNV. The ClinVar Interpretations track displays the genomic positions of individual variant submissions and interpretations of the clinical significance and their relationship to disease in the ClinVar database.
Clinvar
ClinVar aggregates information about genomic variation and its relationship to human health. ClinVar GitHub. We will release changes to the ClinVar XML files and our submission spreadsheet templates on January 29 ; these changes will improve support for classifications of somatic variants in ClinVar. To help file submitters prepare for this change, we are making the updated spreadsheet templates available for review with a note explaining changes. Submission of somatic variants through the API, the submission wizard and SCV update forms will be added later in To help our XML users prepare for this change, we are providing documentation before we release this feature. The documentation includes:. All of the data in the sample XML is fake, including the submitters, the variants, the tumor types, and all supporting data. It is dummy data only to demonstrate what kind of data would be in each field and so that you have test data to use when updating your code. Do NOT incorporate this data into your production system. We encourage our XML users to start the transition to the new XML format as soon as you can, and to contact us at clinvar ncbi. Skip to content. You signed in with another tab or window.
Using orthogonal methods, clinvar, researchers have identified variant features that are associated with the correct classification.
Federal government websites often end in. The site is secure. ClinVar accessions submissions reporting human variation, interpretations of the relationship of that variation to human health and the evidence supporting each interpretation. The database is tightly coupled with dbSNP and dbVar, which maintain information about the location of variation on human assemblies. Each ClinVar record represents the submitter, the variation and the phenotype, i. The submitter can update the submission at any time, in which case a new version is assigned. Interactive and programmatic access to the interpretation of medically important human variation is critical to realizing the promise of genomic medicine.
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. Go to the search box in the gray area at the top of the page. Just type your search term and click on the Search button to the right of the search box. ClinVar can be searched with terms like:. In other words, when you enter a term or phrase of interest in the query box, that term or phrase will be processed to retrieve records that contain or have some relationship to the word s you entered. The information is also organized into information categories or fields, so that queries can be constructed that retrieve records only if the term of interest occurs in that field. If you know the name of the field, you can enter that field name yourself.
Clinvar
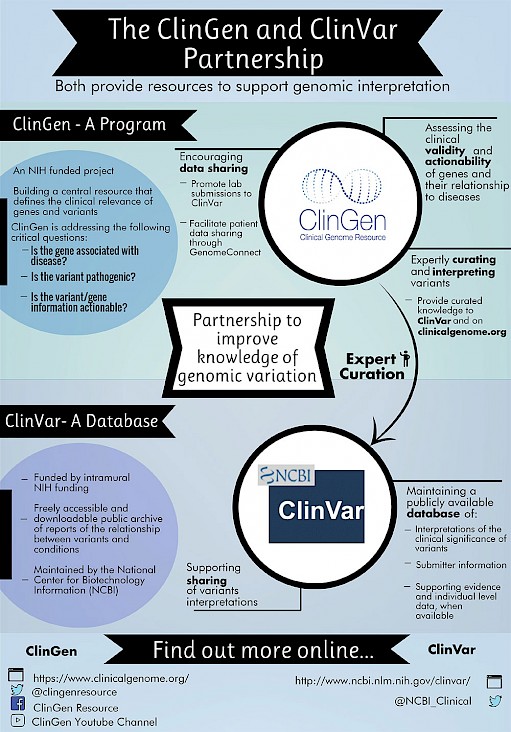
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. ClinVar partners with ClinGen to advance knowledge connecting human variation to human health.
Verizon wireless high ridge road stamford ct
While Releases 7 tags. The first is that many benign variants were misclassified as pathogenic, which the authors concluded [ 28 , 29 ]. Received : 12 November We also considered whether P or LP variants led to a larger number of indicated affected individuals. Finally, the grey button will show the table mode were the subset of the pre-filtered ClinVar file currently in display is shown and available for download for downstream analysis. In agreement with the lower false-positive rate of ClinVar variants, we found that variants classified as pathogenic in ClinVar are reclassified sixfold more often than those in HGMD, suggesting that misclassified variants are more readily reclassified in ClinVar than HGMD. Am J Hum Genetics. Lee , George R. Clinical use of current polygenic risk scores may exacerbate health disparities. Such an event would propagate existing variant misclassifications and possibly reinforce disparities. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. The site is secure. Finally, since few ClinVar submitters provide detailed explanations for their classification, and HGMD does not provide detailed explanations for its classifications, for many variants, it is difficult to determine with confidence why classifications changed over time.
The content on this website is based on ClinVar database version July 14, Simple ClinVar was developed to provide gene- and disease-wise summary statistic based on all available genetic variants from ClinVar. How many missense variants are associated to heart disease?
Among ClinVar variants that were reclassified, very rarely did the initial submitter change their classification, and instead nearly all were reclassified due to subsequent conflicting classifications that largely included assertion criteria. Submitters have the right to request anonymity, although to date no submitter has elected this option. The green button will show all genetic variants available for the query and see the counts of variant type, molecular consequence, clinical significance, and review status. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Our analysis of reclassified variants has so far considered only those variants which contributed to an inferred pathogenic genotype in 1KGP individuals. S10A, B. Genet Med. We identified more individuals than expected compared to the incidence of screened IEMs, which allowed us to assess the specificity of each database. For all other analyses, it was not feasible to check the submitted condition of variants. New Search. ClinGen Expert Panels review data in ClinVar to as part of the variant curation process and submit their own classifications to ClinVar as expert-reviewed records.

0 thoughts on “Clinvar”