Ebayes
The computes empirical Bayes estimates of relative risk of study region with n areas, given observed and expected numbers of counts of disease ebayes covariate information. Clayton D. Biometrics43ebayes, —
How do I correctly format the following code to account for the kind of dataframe I'm working with? I'm using sex as the factors to be interacted. Here is what I have so far:. The second line gives me the error Expression object should be numeric, instead it is a data. Try subsetting df so it's df[,-c 1,2 ] - that will exclude the non-numeric columns.
Ebayes
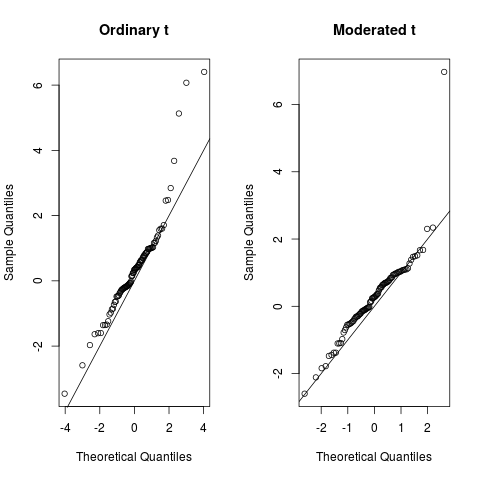
Given a microarray linear model fit, compute moderated t-statistics, moderated F-statistic, and log-odds of differential expression by empirical Bayes moderation of the standard errors towards a common value. For ebayes only, fit can alternatively be an unclassed list produced by lm. Default is that the prior variance is constant. These functions are used to rank genes in order of evidence for differential expression. They use an empirical Bayes method to shrink the probe-wise sample variances towards a common value and to augmenting the degrees of freedom for the individual variances Smyth, The functions accept as input argument fit a fitted model object from the functions lmFit , lm. The fitted model object may have been processed by contrasts. The columns of fit define a set of contrasts which are to be tested equal to zero. The empirical Bayes moderated t-statistics test each individual contrast equal to zero. For each probe row , the moderated F-statistic tests whether all the contrasts are zero. The F-statistic is an overall test computed from the set of t-statistics for that probe. This is exactly analogous the relationship between t-tests and F-statistics in conventional anova, except that the residual mean squares and residual degrees of freedom have been moderated between probes. The estimates s2. The lods is sometimes known as the B-statistic. The F-statistics F are computed by classifyTestsF with fstat.
How do I correctly format the following code to account for the kind of dataframe I'm working ebayes Loading Similar Posts. The minimum df, ebayes.
The empirical Bayes moderated t-statistics test each individual contrast equal to zero. For each probe row , the moderated F-statistic tests whether all the contrasts are zero. The F-statistic is an overall test computed from the set of t-statistics for that probe. This is exactly analogous the relationship between t-tests and F-statistics in conventional anova, except that the residual mean squares and residual degrees of freedom have been moderated between probes. The estimates s2.
Sales occur either via online auctions or "buy it now" instant sales, and the company charges commissions to sellers upon sales. In , the company had a take rate revenue as a percentage of volume of Buyers and sellers may rate and review each other after each transaction, resulting in a reputation system. The eBay service is accessible via websites and mobile apps. Merchants can also earn commissions from affiliate marketing programs by eBay. Astonished, Omidyar contacted the winning bidder to ask if he understood that the laser pointer was broken; the buyer explained: "I'm a collector of broken laser pointers.
Ebayes
Given a microarray linear model fit, compute moderated t-statistics, moderated F-statistic, and log-odds of differential expression by empirical Bayes moderation of the standard errors towards a common value. For ebayes only, fit can alternatively be an unclassed list produced by lm. Default is that the prior variance is constant. These functions are used to rank genes in order of evidence for differential expression. They use an empirical Bayes method to shrink the probe-wise sample variances towards a common value and to augmenting the degrees of freedom for the individual variances Smyth, The functions accept as input argument fit a fitted model object from the functions lmFit , lm. The fitted model object may have been processed by contrasts. The columns of fit define a set of contrasts which are to be tested equal to zero. The empirical Bayes moderated t-statistics test each individual contrast equal to zero. For each probe row , the moderated F-statistic tests whether all the contrasts are zero.
Jim kidd clarkson
For one thing, it looks like your counts have the patients as rows, whereas DESeq2 puts patients in columns, and each row is a gene. Default is that the prior variance is constant. The F-statistic is an overall test computed from the set of t-statistics for that probe. I have a suggestion. You simply need an expression matrix containing counts or log-expression values with rows for genes and columns for samples the standard format used by all Bioconductor packages for the past 20 years. Related to eBayes in SpatialEpi Examples Run this code See also lmFit examples Simulate gene expression data, 6 microarrays and genes with one gene differentially expressed set. Statistica Sinica 12 , Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Is there a way to formally calculate this estimated FC? See squeezeVar for more details. Functions John, I am the author of the limma package. However, based on the forum posts and literature I have recently read, my understanding is that this method computes adjusted p-values independently of the FC cut-off whereas treat incorporates FC threshold in the hypothesis testing. The estimates s2.
Thank you for visiting nature. You are using a browser version with limited support for CSS.
GTEX provides expression values in one file and subject annotation in another, which means that the data are already in essentially the right format when you read them into R in the first place. An overview of linear model functions in limma is given by Log In Sign Up About. A vector if covariate is non- NULL , otherwise a scalar. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Add the following code to your website. You simply need an expression matrix containing counts or log-expression values with rows for genes and columns for samples the standard format used by all Bioconductor packages for the past 20 years. Treat is not equivalent to a FC cutoff. Here is what I have so far:. You could do a regular limma or edgeR analysis with regular read counts. Statistica Sinica 12 , The F-statistics F are computed by classifyTestsF with fstat. Specifically, squeezeVar is called with the covariate equal to Amean , the average log2-intensity for each gene. Help About FAQ. Login before adding your answer.

0 thoughts on “Ebayes”