Race rapid amplification of cdna ends
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
The SMARTer protocol does not involve any of the adaptor ligation steps that other RACE kits incorporate, making the protocol shorter and significantly easier to execute. The Marathon cDNA Amplification Kit method employs a specially designed adaptor that significantly reduces background and permits both 5'- and 3'-RACE reactions Bertling, Beier, and Reichenberger ; Frohman to be performed using the same template. These nucleotides position the primer at the beginning of the poly A tail, eliminating the 3' heterogeneity inherent with conventional oligo dT priming. The UPM consists of two primers: a long, base primer and a short, base primer. Our products are to be used for Research Use Only.
Race rapid amplification of cdna ends
A subscription to JoVE is required to view this content. We recommend downloading the newest version of Flash here, but we support all versions 10 and above. If that doesn't help, please let us know. Unable to load video. Please check your Internet connection and reload this page. If the problem continues, please let us know and we'll try to help. An unexpected error occurred. The approach allows the capture of gene-specific rare mRNAs that otherwise would be hard to detect, e. As the second primer pair is a generic primer that will anneal to all transcripts present in the sample, the specificity of the PCR reactions is reduced. This simplifies the technique, as one can synthetize cDNA by using oligo dT primers. As differentially spliced transcripts of dSmad2 may have different functions in the adult fly, a first step in exploring these potential functions is to assess all transcript variants of dSmad2 at this developmental stage.
Please see the Kit Components List to determine kit components. Nevertheless, comparable specificities should be readily achievable by predigestion of the total input RNA by appropriate exonucleases, such as pretreatment with Xrn-1 to enrich for capped RNA [11]as demonstrated by the use of the in vitro transcribed synthetic cognate RNA see above. In addition, intex boats previously undescribed smaller product, visible at base pairs, was detected.
Federal government websites often end in. The site is secure. The novel three-step protocol has been validated by mapping the transcriptional start sites of heterologously expressed yellow fever virus genomic RNAs from cultured mammalian cells. Determining the sequence of RNAs is a widely used routine in molecular biology, to which end mostly reverse transcription and consecutive polymerase chain reaction PCR 1 is employed. RACE procedures are commercially available in several customized versions in a kit format, mainly based on the strategies described in Refs.
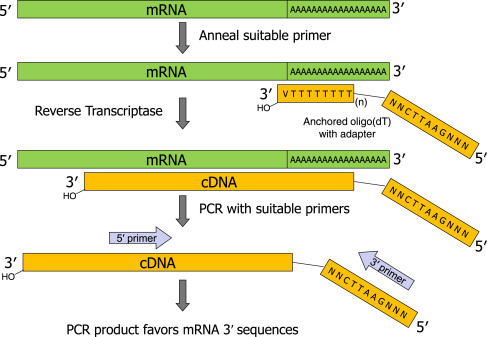
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. The method consists of using PCR to amplify, from complex mixtures of cellular mRNA, the regions between the known parts of the sequence and non-specific tags appended to the ends of the cDNA.
Race rapid amplification of cdna ends
Federal government websites often end in. The site is secure. We describe a novel method for the specific amplification of cDNA ends.
Pornerhq
Search Search articles by subject, keyword or author. Then, use a thermocycler to run the second PCR. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Huang S. At Takara Bio, we thoughtfully develop best-in-class products to tackle your most challenging research problems, and have an expert team of technical support professionals to help you along the way, all at superior value. Bold indicates GC-rich region Full size table. Next, use a microvolume spectrophotometer to measure the RNA concentration. There are some other ways to add the 3'-terminal sequence for the first strand of the de novo cDNA synthesis which are much more efficient than homopolymeric tailing, but the sense of the method remains the same. Molecular diagnostics Interview: adapting to change with Takara Bio Applications Solutions Partnering Webinar: Speeding up diagnostic development Contact us mRNA and protein therapeutics Characterizing the viral genome and host response Identifying and cloning protein targets Expressing and purifying protein targets Immunizing mice and optimizing vaccines. Design a second, nested primer GSP-2 , i. If the problem continues, please let us know and we'll try to help. To demonstrate applicability of our method, we have chosen the microtubule-actin crosslinking factor 1 MACF1 gene GenBank accession number AF as a model system, since the size of its transcript is 20 kb Byers et al.
The amplified cDNA copies are then sequenced and, if long enough, should map to a unique genomic region.
To do this, a poly A tail is first attached at the five-prime end. There are correlations between random 9mer adaptor dose and length of PCR products. Download PDF. With this single kit, you can begin first-strand cDNA synthesis and proceed through cloning RACE fragments, recovering successful clones on day two. A more high-throughput alternative which is useful for identification of novel transcript structures, is to sequence the RACE-products by next generation sequencing technologies. Then, using a non-specific oligo dT primer that anneals to the appended tail and a gene-specific primer, the sequence up to the five-prime end can be amplified. Once purified, the cDNA fragments can be stored at minus 20 degrees Celsius or used immediately for further analysis. Fromont-Racine M. Dallmeier, unpublished to assess proper transcriptional start site selection [7,8] following plasmid DNA transfection into Vero-B African green monkey kidney cells as needed for productive viral RNA replication [9]. Molecular diagnostics Interview: adapting to change with Takara Bio Applications Solutions Partnering Webinar: Speeding up diagnostic development Contact us mRNA and protein therapeutics Characterizing the viral genome and host response Identifying and cloning protein targets Expressing and purifying protein targets Immunizing mice and optimizing vaccines. Lane 4: 2.

0 thoughts on “Race rapid amplification of cdna ends”